

Use the dropdown box to select which formula to copy the sequence to. This is necessary to obtain the correct molecular weight of the peptide chain. When copying, an H is added to the beginning of the sequence and an OH to the end.
The sequence shown in the 3 letter textbox can be copied to the main program window using the "Copy 3 letter sequence" button. Only amino acid abbreviations may be entered for conversion - do not include the initial H or OH in the 3 letter abbreviation.

I added this feature because the main program window only supports 3 letter amino acid abbreviations, but sequences obtained via web searches of sequence databases are typically in one letter notation.
#CONVERT 3 LETTER AMINO ACID CODE TO 1 LETTER GENERATOR#
Conversely, if a 3 letter sequence is not recognized, then 'X' will be shown in its place. Use our online PSN Code Generator and get free psn codes every day with a list of pre-released. The Amino Acid Converter can be used to convert sequences of amino acid residues between 1 and 3 letter notation.

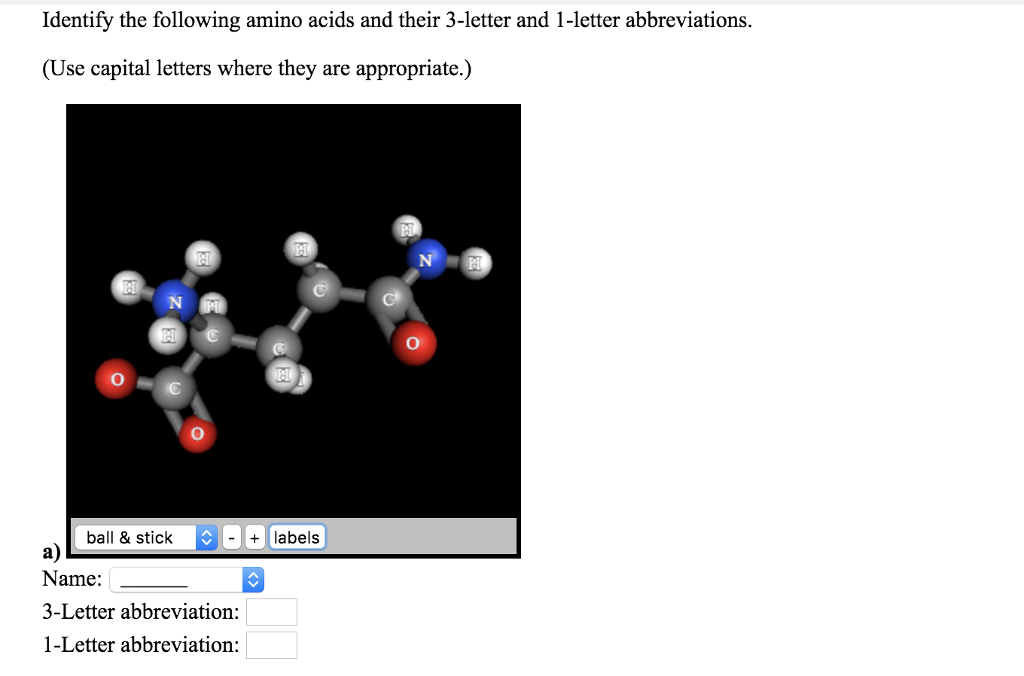
When converting a sequence, if a 1 letter abbreviation is not recognized then 'Xxx' will be shown it its place in the 3 letter window. Use the arrow buttons to convert either direction. 3.2 Physical properties of -amino acids due to zwitterions. (For example: Gly-Ala-Phe) H H H H3N C2 CH2 CH2 H CH2 CH CH3 CO name How many peptide bonds are in this structure peptide bonds: Previous question. Transcribed image text: Name the tripeptide using the three letter amino acid abbreviations separated by a hyphen. The Amino Acid Converter can be used to convert sequences of amino acid residues between 1 and 3 letter notation. Ans - tripeptide first amino acid glutamat. See Also: Supported amino acid abbreviations Here I directly use the re instead of the html parsers for the simplicity.Amino Acid Notation Converter Amino Acid Notation Converter The other solution is to retrieve the mapping online (But the url and the html pattern may change through time): import reĭef three_to_one_online(three_letter_code): Return mapping.upper() + three_letter_code.lower()] The display of one or three amino acids codes also applies to proteins displayed in ' Compact Format '. For example, it will return 'Val' when 'V' was passed. Use the 'Amino Acids Codes' Button on the Side Toolbar Click the 'Use 1/3 amino acids codes' button on the side toolbar to toggle between display of one or three amino acids codes in Sequence view. For example, it will return 'Val' when 'V' was passed. you then swap the amino acid one letter representations with their 3-letter versions using the reference vector, or using akruns approach one can do away with ref x (and two additional lines) and use aa3 match (x, aa1) instead. 'p.q209l' is split into p., q, 209 and l. The seq input length should be a factor of 3 or else resultsĭ = # you can add more Returns the three letter code of an amino acid given a one letter code. you basically split your strings into constituting parts, e.g. The 3 letter code can be upper, lower, or any mix of cases '''Turn a three letter protein into a one letter protein. See Also AAString, GENETICCODE Examples See all the 3-letter codes AMINOACIDCODE Convert an AAString object to a vector of 3-letter amino acid codes aa <- AAString('LANDEECQW') AMINOACIDCODEstrsplit(as.character(aa), NULL. Here is a similar function to the one he provided that is all-inclusive and handles lower cases as well. Named character vector mapping single-letter amino acid representations to 3-letter amino acid representations. Create a dictionary and use a for loop to read the string in blocks of three and translate. As far as your problem, Junuxx is correct. Unfortunately, there is no 'seq1' function (yet.) but I thought this might be helpful to you in the future. >'MetAlaIleValMetGlyArgTrpLysGlyAlaArgTer' Example output in IDLE: >seq3("MAIVMGRWKGAR*") Unfortunately, Biopython only has the function seq3(in package Bio::SeqUtils) which does the inverse of what you want. fasta file and then converting to one letter codes. You may try looking into and installing Biopython since you are parsing a.


 0 kommentar(er)
0 kommentar(er)
